Molecular Biology teaching strategy for In Silicogeneediting: A disruptive experience
DOI:
https://doi.org/10.26423/rcpi.v11i1.684Keywords:
DNA, project-based learning, biotechnology, higher educationAbstract
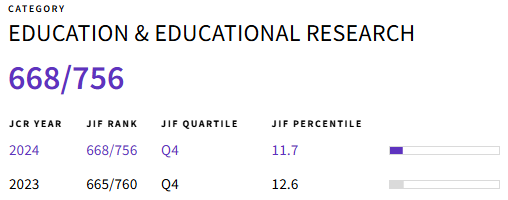
In silico techniques are used to simulate experiments using molecular biology computational tools. This study aimed to promote the use of in silico assays in students of biotechnology engineering at the Universidad Regional Amazónica Ikiam. The methodology consisted of three phases: a) planning and organization, b) practice and execution, and c) project evaluation. In this sense, each group used bibliographic sources indexed in Scopus, Springer, and PubMed; databases such as Bioweb and Genbank; genome banks AddGene, EMBL, and NCBI; to assemble a new plasmid in Benchling. The main result was six projects that sought alternatives to current health, environmental, and agricultural challenges. Among the projects linked to health, there were two projects, G-1 and G-2, while the projects focused on the environmental component, G-4 and G-5, and those related to agricultural improvement, G-3, and G-6. According to the survey conducted at the end of the semester, the classroom projects were highly accepted. It is suggested that these strategies be utilized when studying topics associated with the biological sciences.
Downloads
References
Alonso-Fernández, Cristina; Calvo-Morata, Antonio; Freire, Manuel; Martínez-Ortiz, Iván; y Fernández-Manjón, Baltasar (2019). Applications of data science to game learning analytics data: A systematic literature review. Computers & Education, 141(2019), 103612. https://doi.org/10.1016/j.compedu.2019.103612
Antunes, Luisa (6 junio 2022). Genome Editing in Humans: A survey of law, regulation and governance principles. By Scientific Foresight (STOA). EPRS | European Parliamentary Research Service. Disponible en: https://epthinktank.eu/2022/06/06/genome-editing-in-humans-a-survey-of-law-regulation-and-governance-principles/
Aukema, Kelly G.; Escalante, Diego E.; Maltby, Meghan M.; Bera, Asim K.; Aksan, Alptekin; y Wackett, Lawrence P. (2017). In Silico Identification of Bioremediation Potential: Carbamazepine and Other Recalcitrant Personal Care Products. Environmental Science & Technology, 51(2), 880-888. https://doi.org/10.1021/acs.est.6b04345
Bhat, Gh Rasool; Sethi, Itty; Rah, Bilal; Kumar, Rakesh; y Afroze, Dil (2022). Innovative in Silico Approaches for Characterization of Genes and Proteins. Frontiers in Genetics, 13(2022), 1-20. https://doi.org/10.3389/fgene.2022.865182
Capa Gaona Jhoana; Ordoñez Mendoza Rubí; Chicaiza Cristhian Ortiz; Loján María del Cisne; Alvarado Ávila Ginno; y Romero Paguay José. (2022). Enseñanza de Química, Bioquímica y Biotecnología en tiempos de pandemia: herramientas y experiencias. Zenodo. https://doi.org/10.5281/zenodo.6582193
Capecchi, Mario R. (1989). Altering the Genome by Homologous Recombination. Science, 244(4910), 1288-1292. https://doi.org/10.1126/science.2660260
Carbonell, Pablo; Le Feuvre, Rosalind; Takano, Eriko; y Scrutton, Nigel S. (2020). In silico design and automated learning to boost next-generation smart biomanufacturing. Synthetic Biology (Oxford, England), 5(1), ysaa020. https://doi.org/10.1093/synbio/ysaa020
Chicaiza-Ortiz, Cristhian D.; Rivadeneira-Arias, Virginia del C.; Herrera-Feijoo, Robinson J.; y Andrade, Jean Carlo (2023). Biotecnología Ambiental, Aplicaciones y Tendencias. Editorial Grupo AEA. Disponible en: https://doi.org/10.55813/egaea.l.2022.25
Clement, Kendell; Hsu, J. Yonathan; Canver, Matthew C.; Joung, J. Keith; y Pinello, Luca (2020). Technologies and Computational Analysis Strategies for CRISPR Applications. Molecular Cell, 79, 11-29. https://doi.org/https://doi.org/10.1016/j.molcel.2020.06.012
Coronas-Serna, Julia M.; Del Val, Elba; Kagan, Jonathan C.; Molina, María; y Cid, Victor J. (2021). Heterologous Expression and Assembly of Human TLR Signaling Components in Saccharomyces cerevisiae. Biomolecules, 11(11), 1737. Disponible en: https://doi.org/10.3390/biom11111737
Correa, B., Rios, L., Volpi e Silva, N., Prado, G. S., & Lopes, J. H. (2021). Technology in Plant Genome Editing. Embrapa.
Cortes-Hernandez, Paulina; y Domínguez-Ramírez, Lenin (2019). In Silico Mutagenesis, Docking, and Molecular Dynamics: Their Role in Biosensor Design for Environmental Analysis and Monitoring BT - Ecopharmacovigilance: Multidisciplinary Approaches to Environmental Safety of Medicines (L. M. Gómez-Oliván (ed.); pp. 221-234). Springer International Publishing. https://doi.org/10.1007/698_2017_144
El-Bassyouni, H.; y Mohammed, M. (2018). Genome Editing: A Review of Literature. LAP LAMBERT Academic Publishing.
Estrellado, R. A., Freer; E. A., Motsipak, J.; Rosenberg, J. M.; y Velásquez, I. C. (2020). Data science in education using R. London, England: Routledge. Nb. All authors contributed equally.
Furtado, Rafael Nogueira (2019). Gene editing: the risks and benefits of modifying human DNA. Rev. Bioét., 27(2), 223-233. https://doi.org/10.1590/1983-80422019272304
García-Carpintero, E. (2017). El portafolio como metodología de enseñanza-aprendizaje y evaluación en el practicum: percepciones de los estudiantes. REDU. Revista de Docencia Universitaria, 15(1), 241-257. Disponible en: https://doi.org/10.4995/redu.2017.6043
Garcillán-Barcia, M. Pilar; Pluta, Radoslaw; Lorenzo-Díaz, Fabián; Bravo, Alicia; y Espinosa, Manuel (2022). The Facts and Family Secrets of Plasmids That Replicate via the Rolling-Circle Mechanism. Microbiology and molecular biology reviews: MMBR, 86(1), e0022220. https://doi.org/10.1128/MMBR.00222-20
Karre, Ashok (2020). Gene editing technology, Information Technology. Disponible en: https://www.researchgate.net/profile/Ashok-Karre/publication/347442835_GENE_EDITING_TECHNOLOGY/links/5fdc3550299bf140881b5828/GENE-EDITING-TECHNOLOGY.pdf
Khalil, Ahmad M. (2020). The genome editing revolution: review. Journal of Genetic Engineering and Biotechnology, 68(2020), 68. https://doi.org/10.1186/s43141-020-00078-y
Khan, Sikandar Hayat (2019). Genome-Editing Technologies: Concept, Pros, and Cons of Various Genome-Editing Techniques and Bioethical Concerns for Clinical Application. Molecular Therapy - Nucleic Acids, 16(June), 326-334. Disponible en: https://doi.org/10.1016/j.omtn.2019.02.027
Khurshed, Mohammed; Molenaar, Remco J.; y Van Noorden, Cornelis JF (2019). A simple in silico approach to generate gene-expression profiles from subsets of cancer genomics data. BioTechniques, 67(4), 172-176. https://doi.org/10.2144/btn-2018-0179
Labun, Kornel (2020). In silico design and analysis of targeted genome editing with CRISPR [Tesis PhD]. University of Bergen, Noruega. Disponible en: https://bora.uib.no/bora-xmlui/handle/1956/21443
Moradi, Mohammad; Golmohammadi, Reza; Najafi, Ali; Moosazadeh Moghaddam, Mehrdad; Fasihi-Ramandi, Mahdi; y Mirnejad, Reza (2022). A contemporary review on the important role of in silico approaches for managing different aspects of COVID-19 crisis. Informatics in medicine unlocked, 28(100862), 2352-9148. Disponible en: https://doi.org/https://doi.org/10.1016/j.imu.2022.100862
Murray, David; Doran, Peter; MacMathuna, Padraic; y Moss, Alan C. (2007). In silico gene expression analysis – an overview. Molecular Cancer, 50(2007), 6-50. Disponible en: https://doi.org/10.1186/1476-4598-6-50
Pradhan, Chandan Kumar; Nayak, Suraja, Kumar; y Baliyarsingh, Bighneswar (2022). In Silico Tools and Approach of CRISPR Application in Agriculture. Advances in Agricultural and Industrial Microbiology, 2(2022), 177-189. https://doi.org/10.1007/978-981-16-9682-4_10
Pratami, Mentari Putri; Fendiyanto, Miftahul Huda; Satrio, Rizky Dwi; Nikmah, Isna Arofatun; Awwanah, Mo; Farah, Nadya; Permata Sari, Nastiti Intan; y Nurhadiyanta (2022). In-silico Genome Editing Identification and Functional Protein Change of Chlamydomonas reinhardtii Acetyl-CoA Carboxylase (CrACCase). Jordan Journal of Biological Sciences, 15(3), 431 – 440. Disponible en: https://doi.org/https://doi.org/10.54319/jjbs/150312
Rahnama, Hassan; Nikmard, Mahdi; Abolhasani, Mohsen; Osfoori, Rahim, Sanjarian, Forough; y Habashi, Ali Akbar (2017). Immune analysis of cry1Ab-genetically modified potato by in-silico analysis and animal model. Food Science and Biotechnology, 26(5), 1437-1445. Disponible en: https://doi.org/10.1007/s10068-017-0181-4
Sandoval Rodríguez, Ana S.; Mena Enríquez, Mayra; y Márquez Aguirre, Aana L. (2013). Capítulo13: Vectores de clonación y expresión. En A. M. Salazar Montes, A. S. Sandoval Rodríguez, & J. S. Armendáriz Borunda (Eds.), Biología molecular. Fundamentos y aplicaciones en las ciencias de la salud. McGraw-Hill Education. Disponible en: http://accessmedicina.mhmedical.com/content.aspx?aid=1118679813
Silva, E. A. J., Estevam, E. B. B., Silva, T. S., Nicolella, H. D., Furtado, R. A., Alves, C. C. F., Souchie, E. L., Martins, C. H. G., Tavares, D. C., Barbosa, L. C. A., y Miranda, M. L. D. (2019). Antibacterial and antiproliferative activities of the fresh leaf essential oil of Psidium guajava L. (Myrtaceae). Braz J Biol, 79(4), 697-702. Disponible en: https://doi.org/10.1590/1519-6984.189089
Taldaev, Amir; Terekhov, Roman; Nikitin, Ilya; Zhevlakova, Anastasiya; y Selivanova, Irina (2022). Insights into the Pharmacological Effects of Flavonoids: The Systematic Review of Computer Modeling. International Journal of Molecular Sciences, 23(11). Disponible en: https://doi.org/10.3390/ijms23116023
Tang, Jinling; Lee, Trevor; y Sun Tao (2019). Single-nucleotide editing: From principle, optimization to application. Human Mutation 40(12) 2171-2183. Disponible en: https://doi.org/10.1002/humu.23819
Teixeira Gomes, Ana F.; Fortunado de Medeiros, Wendjilla; Silva de Oliveira, Gerciane; Medeiros, Isaiane; Da Silva Maia, Juliana K.; Leal Bezerra, Ingrid W.; Piuvezam, Grasiela; y Heloneida de Araujo Morais, Ana (2022). In silico structure-based designers of therapeutic targets for diabetes mellitus or obesity: A protocol for systematic review. PLOS ONE, 17(12), e0279039.
Timo, Giulia O.; Reis, Rodrigo, S. S. V. D.; Melo, Adriana F.; Costa, Thales V. L.; Magalhães, Pérola O.; y Homem-de-Mello, Mauricio (2019). Predictive Power of In Silico Approach to Evaluate Chemicals against M. tuberculosis: A Systematic Review. Pharmaceuticals (Basel, Switzerland), 12(3), 1424-8247. Disponible en: https://doi.org/https://doi.org/10.3390/ph12030135
Todke, Pooja A.; y Devarajan, Padma V. (2022). In-silico approach as a tool for selection of excipients for safer amphotericin B nanoformulations. Journal of Controlled Release, 349(2022), 756-764. Disponible en: https://doi.org/https://doi.org/10.1016/j.jconrel.2022.07.030
Trump, Benjamin; Cummings, Christopher; Klasa, Kasia; Galaitsi, Stephanie; y Linkov, Igor (2022). Governing biotechnology to provide safety and security and address ethical, legal, and social implications. Frontiers in Genetics, 13, 1052371. https://doi.org/10.3389/fgene.2022.1052371
Zhang, Yuwei; Zhao, Guofang; Ahmed, Hadi Fatma; Yi, Tianfei; Hu, Shiyun; Cai, Ting; y Liao, Qi (2020). In silico Method in CRISPR/Cas System: An Expedite and Powerful Booster. Frontiers in Oncology, 10(2020), 1-20. Disponible en: https://doi.org/10.3389/fonc.2020.58440
Downloads
Published
Issue
Section
License
El titular de los derechos de autor de la obra, otorga derechos de uso a los lectores mediante la licencia Creative Commons Atribución-NoComercial-CompartirIgual 4.0 Internacional. Esto permite el acceso gratuito inmediato a la obra y permite a cualquier usuario leer, descargar, copiar, distribuir, imprimir, buscar o vincular a los textos completos de los artículos, rastrearlos para su indexación, pasarlos como datos al software o usarlos para cualquier otro propósito legal.
Cuando la obra es aprobada y aceptada para su publicación, los autores conservan los derechos de autor sin restricciones, cediendo únicamente los derechos de reproducción, distribución para su explotación en formato de papel, así como en cualquier otro soporte magnético, óptico y digital.